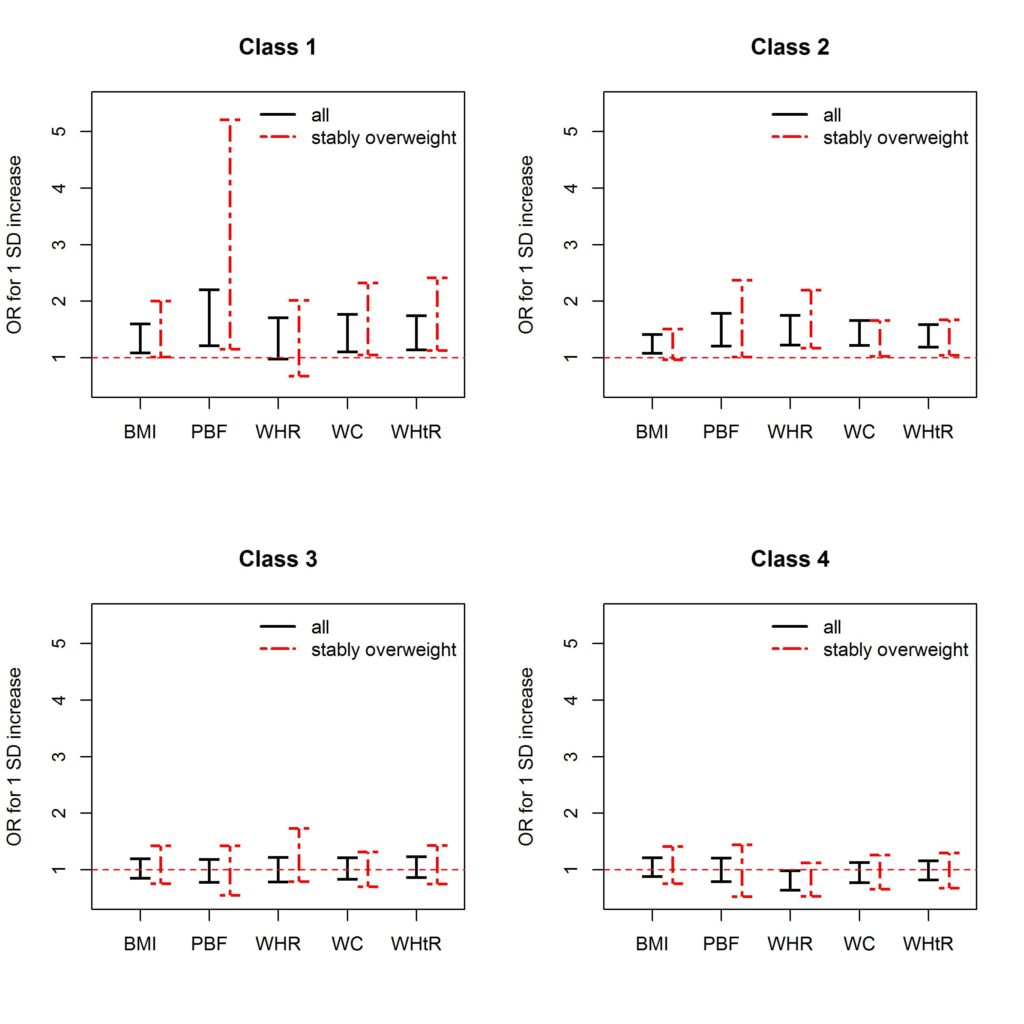
This is an example of how to visualize odds ratio using generic plotting functions in R, e.g. functions from {base} and {graphics} packages.
By applying multinomial regression using R package {nnet}, odds ratio was calculated separately for asthma classes and for different obesity metrics. And the same model was fitted restricted to stably overweight participants as a sensitivity analysis. Odds ratios for different obesity metrics were plotted side-by-side for comparison, and the odds ratios from the main model and from the sensitivity analysis were distinguished by color and line type. See the original publication for details.
### Not to run ###
library(nnet)
# multinomial regression on different obesity measures
model <-
'lc.w.ref ~ age_s3 + sex_s1 + educ_cum + smok_cum_s3 + area_s3 + '
obesity <- c('bmi_s3', 'pbf_kyle_s3', 'waist_hip_ratio_s3', 'waist_s3',
'waist_height_ratio_s3')
tab.all <- list()
for(obe in obesity){
test.model <- as.formula(paste0(model, obe))
test <- multinom(test.model, data = subd)
# calculate the p values manually
z.values <- summary(test)$coefficients/summary(test)$standard.errors
p.values <- pnorm(abs(z.values), 0, 1, lower.tail = FALSE) * 2
# produce the coefficient table for each class
tab.all[[obe]] <- list()
for(class in rownames(coef(test))){
tab.all[[obe]][[class]] <-
data.frame(beta = coef(test)[class, ],
se = summary(test)$standard.errors[class, ],
p.value = p.values[class, ])
}
}
# multinomial regression on different obesity measures,
# restricted to the stably overweight
model <-
'lc.w.ref ~ age_s3 + sex_s1 + educ_cum + smok_cum_s3 + area_s3 + '
obesity <- c('bmi_s3', 'pbf_kyle_s3', 'waist_hip_ratio_s3', 'waist_s3',
'waist_height_ratio_s3')
tab.stab <- list()
for(obe in obesity){
test.model <- as.formula(paste0(model, obe))
test <- multinom(test.model, data = subd)
# calculate the p values manually
z.values <- summary(test)$coefficients/summary(test)$standard.errors
p.values <- pnorm(abs(z.values), 0, 1, lower.tail = FALSE) * 2
# produce the coefficient table for each class
tab.stab[[obe]] <- list()
for(class in rownames(coef(test))){
tab.stab[[obe]][[class]] <-
data.frame(beta = coef(test)[class, ],
se = summary(test)$standard.errors[class, ],
p.value = p.values[class, ])
}
}
# compute the odds ratios
or.all <- list()
for(obe in names(tab.all)){
or.all[[obe]] <- data.frame(OR = NULL, lower = NULL, upper = NULL)
for(class in names(tab.all[[obe]])){
beta <- tab.all[[obe]][[class]][obe, 'beta']
se <- tab.all[[obe]][[class]][obe, 'se']
or.all[[obe]][class, 'OR'] <- exp(beta)
or.all[[obe]][class, 'lower'] <- exp(beta - qnorm(0.975) * se)
or.all[[obe]][class, 'upper'] <- exp(beta + qnorm(0.975) * se)
}
}
or.stab <- list()
for(obe in names(tab.stab)){
or.stab[[obe]] <- data.frame(OR = NULL, lower = NULL, upper = NULL)
for(class in names(tab.stab[[obe]])){
beta <- tab.stab[[obe]][[class]][obe, 'beta']
se <- tab.stab[[obe]][[class]][obe, 'se']
or.stab[[obe]][class, 'OR'] <- exp(beta)
or.stab[[obe]][class, 'lower'] <- exp(beta - qnorm(0.975) * se)
or.stab[[obe]][class, 'upper'] <- exp(beta + qnorm(0.975) * se)
}
}
# visualize odds ratios with 95% confidence interval
jpeg('results/figure2_odds_ratio.jpg', width = 3000, height = 3000, res = 400)
layout(matrix(1:4, 2, 2, byrow = TRUE))
for(i in 1:4){
lower.all <- unlist(lapply(or.all, function(x)
x[paste0('class', i), 'lower']))
upper.all <- unlist(lapply(or.all, function(x)
x[paste0('class', i), 'upper']))
lower.stab <- unlist(lapply(or.stab, function(x)
x[paste0('class', i), 'lower']))
upper.stab <- unlist(lapply(or.stab, function(x)
x[paste0('class', i), 'upper']))
plot(0, type = 'n', xlim = c(0.5, 5.5), ylim = c(0.5, 5.5),
ann = FALSE, xaxt = 'n')
title(main = paste('Class', i), ylab = 'OR for 1 SD increase')
axis(1, at = 1:5, labels = c('BMI', 'PBF', 'WHR', 'WC', 'WHtR'))
arrows(1:5, lower.all, 1:5, upper.all,
code = 3, angle = 90, length = 0.075, lwd = 2, lend = 'butt')
arrows(1:5+.3, lower.stab, 1:5+.3, upper.stab,
code = 3, angle = 90, length = 0.075, lwd = 2, lend = 'butt',
lty = 6, col = 2)
abline(h = 1, lty = 2, col = 2)
legend('topright', legend = c('all', 'stably overweight'),
bty = 'n', lty = c(1, 6), lwd = 2, col = 1:2)
}
dev.off()